Spectrogram¶
Description¶
Computes the Spectrogram of 1D signals.
The spectrogram of signal is computed by dividing the signal into \(N_b\) overlapping blocks of the same length and performing the Fourier transform on each block. This allows us to study the changes of the spectra as a function of time.
Note
Dependency on matplotlib.mlab
Parameters¶
- base:
Base The input base. The base can contain many zones, typically, many probes. Each zone must contain the variable(s) on which to compute the spectrogram and the time vector.
- base:
- resize_time_factor: float, default= 1.0
Factor to re-dimensionalize the time variable (time used will be time_var * resize_time_factor). This is useful when you only have the iteration variables and when the time-step used is constant.
- variables: list(str)
The variables that serve as a basis for the spectrogram. The first variable must be the variable representing time.
- time_t0: float, default= None
Time from which the analysis will start. If
None, the analysis will start from the beginning of the time array.
- time_tf: float, default= None
Time at which the analysis will end. If :python`None`, the analysis will go until the end of the time array.
- nb_block: int or float, default= 1
Number of blocks
- pad: int or float, default= 1
Padding of the signal with zeros. pad =1 means that there is no padding. Otherwise, if the length of the signal is Ls, then the length of the padded signal will be Ls*pad.
- overlap: int or float, default= 0.33
The percent of the length of the signal to be overlapped.
- window: str or function, default= ‘hanning’
The window function to be applied to each segment. The possible values are: ‘hanning’, ‘none’, ‘blackman’, ‘hamming’, ‘bartlett’.
If none of the above suits you, you can also set your own custom window function. The window function must be a function that accepts a numpy array representing the signal and it returns a single numpy array representing the windowed signal.
Example : The following code defines a triangular window using the function
triangin thescipy.signallibraryimport scipy.signal def my_triangular_window(signal): return signal * scipy.signal.triang(len(signal)) tt = antares.Treatment('specgram') tt['window'] = my_triangular_window
- scale_by_freq: bool or None, default= None
If the computed spectrogram should be scaled by the sampling frequency. If True the unit of the results will be \(\propto \text{Hz}^{-1}\).
- t_and_f_in_attrs: bool, defaut= True
If
True, the time and frequency vectors will be saved as 1D vectors in the attributes of the instant. IfFalse, they will be saved as 2D matrices resulting from thenumpy.meshgrid(time, freq).
- mode: str, default= ‘psd’
What sort of spectrum to use:
'psd'returns the power spectral density'complex'returns the complex-valued frequency spectrum'magnitude'returns the magnitude spectrum'angle'returns the phase spectrum without unwrapping'phase'returns the phase spectrum with unwrapping
Preconditions¶
Each instant must contain all the variables listed in
variables.
Postconditions¶
The output base contains as many zones as the input base. Each zone contains the same number of instant found in the zone of the input base. Each instant contains the variables on which the spectrogram was calculated as a matrix of dimensions \((N_b \times N_f)\), where \(N_b\) is the number of blocks and \(N_f\) is the number of frequencies that can be captured by the Discrete Fourier Transform.
If t_and_f_in_attrs = True the attributes of the instant
contain the 'time' and 'frequency’ variables as 1D vectors
of length \(N_b\) and \(N_f\). If t_and_f_in_attrs =
False, the 'time' and 'frequency’ are stored as
2D matrices of dimensions \((N_b \times N_f)\).
Main functions¶
- class antares.treatment.TreatmentSpecgram.TreatmentSpecgram¶
- execute()¶
Compute the Power Spectral Density.
- Returns:
A base with the same number of zone and instants as the input base and the spectrogram of the
variables. Time and frequency can be found either in the attributes of the instant or on the instant’s variables depending on the value oft_and_f_in_attrs.- Return type:
Example¶
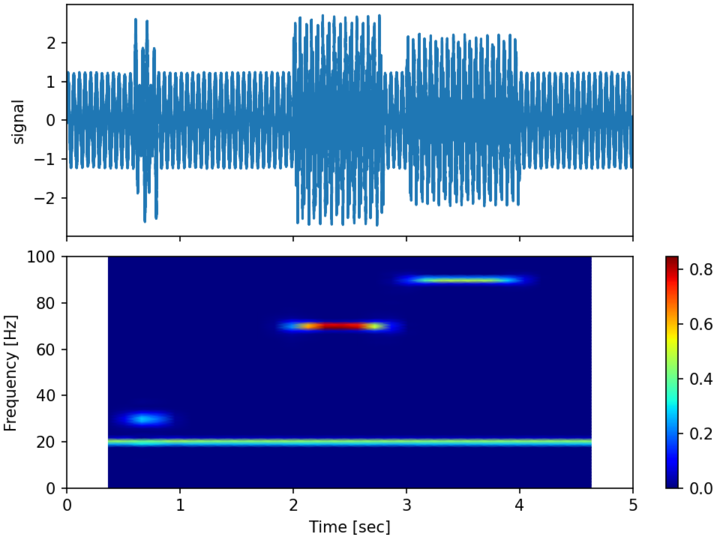
The following example computes the spectrogram of the following 1D signal between \(0 \leq t \leq 5\) with a sampling rate of 10kHz :
A random uniform noise of amplitude 0.25 is also added to the signal.
The spectrogram is computed using 30 blocks with an overlap of 80% and no zero-padding. The Hanning window function is used.
import os
import matplotlib.pyplot as plt
import numpy as np
from antares.api import Base
from antares.treatment import Treatment
np.random.seed(19680801)
output_folder = os.path.join(os.getcwd(), 'OUTPUT', 'SPECGRAM')
os.makedirs(output_folder, exist_ok=True)
## Create signal
## =============
dt = 0.0001
time = np.arange(0.0, 5.0, dt)
s1 = 1 * np.sin(2 * np.pi * 20 * time)
s2 = 1.5 * np.sin(2 * np.pi * 30 * time)
s3 = 1.5 * np.sin(2 * np.pi * 70 * time)
s4 = 1.0 * np.sin(2 * np.pi * 90 * time)
# create a transient "chirp"
s2[time < .6] = s2[0.8 <= time] = 0
s3[time < 2.0] = s3[2.8 <= time] = 0
s4[time < 3.0] = s4[4.0 <= time] = 0
# add some noise into the mix
noise = 0.25 * np.random.uniform(-1, 1, size=len(time))
# The signal
signal = s1 + s2 + s3 + s4 + noise
base = Base()
base.init()
base[0][0]['time'] = time
base[0][0]['signal'] = signal
## Spectrogram treatment
## =====================
treatment = Treatment('specgram')
treatment['base'] = base
treatment['variables'] = ['time', 'signal']
treatment['nb_block'] = 30
treatment['pad'] = 1
treatment['overlap'] = 0.8
treatment['t_and_f_in_attrs'] = True
treatment['window'] = 'hanning'
result = treatment.execute()
t = result[0][0].attrs['time']
f = result[0][0].attrs['frequency']
Pxx = result[0][0]['signal']
## Plot spectrogram
## ===============
jet_colormap = plt.cm.get_cmap("jet")
fig, ax = plt.subplots(nrows=2, sharex=True, constrained_layout=True)
_ = ax[0].plot(time, signal)
p = ax[1].pcolormesh(t, f, Pxx, cmap=jet_colormap, shading='gouraud')
plt.xlabel('Time [sec]')
ax[0].set_ylabel('signal')
ax[1].set_ylabel('Frequency [Hz]')
ax[1].set_ylim([0, 100])
ax[1].set_xlim([0, 5])
fig.colorbar(p, ax=ax[1], orientation='vertical')
plt.savefig(os.path.join(output_folder, 'specgram_output.png'), dpi=150)